Parametric regression models for censored data
Jan Vávra
Exercise 1 (7th November 2022)
Lecture revision: blackboard notes.
PBC (Primary Biliary Cirrhosis) dataset
This data is from the Mayo Clinic trial in primary biliary cirrhosis (PBC) of the liver conducted between 1974 and 1984. A total of 424 PBC patients, referred to Mayo Clinic during that ten-year interval, met eligibility criteria for the randomized placebo controlled trial of the drug D-penicillamine. The first 312 cases in the data set participated in the randomized trial and contain largely complete data. The additional 112 cases did not participate in the clinical trial, but consented to have basic measurements recorded and to be followed for survival. Six of those cases were lost to follow-up shortly after diagnosis, so the data here are on an additional 106 cases as well as the 312 randomized participants.
library(survival)
dim(pbc)## [1] 418 25pbc <- pbc[, c("id", "time", "status", "trt", "sex", "edema", "bili", "albumin", "platelet")]
# id = case number
# time = number of days between registration and the earlier of death, transplantion, or study analysis in July, 1986
# status = status at endpoint, 0/1/2 for censored, transplant, dead
# trt = 1/2/NA for D-penicillamin, placebo, not randomised
# sex = m/f
# edema = 0 no edema, 0.5 untreated or successfully treated, 1 edema despite diuretic therapy
# bili = serum bilirunbin (mg/dl)
# albumin = serum albumin (g/dl)
# platelet = platelet count
head(pbc)## id time status trt sex edema bili albumin platelet
## 1 1 400 2 1 f 1.0 14.5 2.60 190
## 2 2 4500 0 1 f 0.0 1.1 4.14 221
## 3 3 1012 2 1 m 0.5 1.4 3.48 151
## 4 4 1925 2 1 f 0.5 1.8 2.54 183
## 5 5 1504 1 2 f 0.0 3.4 3.53 136
## 6 6 2503 2 2 f 0.0 0.8 3.98 NAsummary(pbc)## id time status trt sex edema bili
## Min. : 1.0 Min. : 41 Min. :0.0000 Min. :1.000 m: 44 Min. :0.0000 Min. : 0.300
## 1st Qu.:105.2 1st Qu.:1093 1st Qu.:0.0000 1st Qu.:1.000 f:374 1st Qu.:0.0000 1st Qu.: 0.800
## Median :209.5 Median :1730 Median :0.0000 Median :1.000 Median :0.0000 Median : 1.400
## Mean :209.5 Mean :1918 Mean :0.8301 Mean :1.494 Mean :0.1005 Mean : 3.221
## 3rd Qu.:313.8 3rd Qu.:2614 3rd Qu.:2.0000 3rd Qu.:2.000 3rd Qu.:0.0000 3rd Qu.: 3.400
## Max. :418.0 Max. :4795 Max. :2.0000 Max. :2.000 Max. :1.0000 Max. :28.000
## NA's :106
## albumin platelet
## Min. :1.960 Min. : 62.0
## 1st Qu.:3.243 1st Qu.:188.5
## Median :3.530 Median :251.0
## Mean :3.497 Mean :257.0
## 3rd Qu.:3.770 3rd Qu.:318.0
## Max. :4.640 Max. :721.0
## NA's :11Exponential regression with arbitrary random censoring
Let us suppose the following model. We are interested in the
distribution of survival time of a patient denoted by \(T\). \(C\)
will denote the (censoring) time when the patient had (or would have
had) transplantation or the time for which he/she has been observed.
What we actually measure is the time of the event, which comes first,
i.e. \(X = \min \{T, C \}\) (variable
time). We also know \(\delta\) - the indicator of what event
actually happened (variable status). Our data consist of
\(n\) = 418 triplets \(\left(X_i, \delta_i, \mathbf{Z}_i\right)\),
where \(\mathbf{Z}_i\) are other
measured covariates.
We suppose that these triplets are stochastically independent of each other, censoring times \(C_i\) are arbitrary and independent of survival times \(T_i\), however, we suppose that \[T_i | \mathbf{Z}_i \sim \mathsf{Exp} \left(\lambda(\mathbf{Z}_i, \boldsymbol\beta) \right), \qquad \text{where} \; \lambda(\mathbf{Z}, \boldsymbol \beta) = \lambda_0 \exp \left\{ \boldsymbol\beta^{\mathsf{T}} \mathbf{Z} \right\} \] are individual hazards arising from linear combination of given covariates. \(\lambda_0\) is called the baseline hazard (\(\lambda = \lambda_0 \Leftrightarrow \mathbf{Z}_i = \mathbf{0}\)). For identifiability purposes we do not allow intercept among covariates \(\mathbf{Z}_i\) (in this notation). The goal is to estimate unknown coefficients \(\boldsymbol \beta\) in order to describe the influence of covariates on the hazard.
We can fit this model in R using glm function with
appropriate setting (see lecture notes).
Usage of glm function
The following must be done:
- Response variable must be set to \(\delta\) indicator.
- Choose appropriate set of covariates (intercept is automatically
included).
- Intercept term corresponds to \(\log \lambda_0\).
- Add
+ offset(log(time))into the model formula. - Set
family = poisson(with canonical \(\log\) link).
However, even if we proceed according to these rules, the following model is still invalid:
fit_cens_WRONG <- glm(status ~ trt + sex + edema + bili + albumin + platelet + offset(log(time)), family=poisson, data = pbc)
summary(fit_cens_WRONG)##
## Call:
## glm(formula = status ~ trt + sex + edema + bili + albumin + platelet +
## offset(log(time)), family = poisson, data = pbc)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.7052 -1.1500 -0.8297 1.0742 3.7721
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -3.7767641 0.5943777 -6.354 2.10e-10 ***
## trt -0.1222921 0.1241319 -0.985 0.324536
## sexf -0.7083493 0.1657342 -4.274 1.92e-05 ***
## edema 0.8306849 0.2182798 3.806 0.000141 ***
## bili 0.0982134 0.0098939 9.927 < 2e-16 ***
## albumin -0.9490988 0.1564953 -6.065 1.32e-09 ***
## platelet -0.0010431 0.0006906 -1.511 0.130910
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for poisson family taken to be 1)
##
## Null deviance: 776.37 on 307 degrees of freedom
## Residual deviance: 533.22 on 301 degrees of freedom
## (110 observations deleted due to missingness)
## AIC: 909.32
##
## Number of Fisher Scoring iterations: 6Find out what is wrong with this model and make necessary changes.
After needed changes we get the following results:
fit_cens <- glm(delta ~ ftrt + fsex + fedema + bili + albumin + platelet + offset(log(time)), family=poisson, data = subdata)
summary(fit_cens)##
## Call:
## glm(formula = delta ~ ftrt + fsex + fedema + bili + albumin +
## platelet + offset(log(time)), family = poisson, data = subdata)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.9482 -0.7927 -0.5976 0.7868 2.6869
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -5.1171852 0.7756655 -6.597 4.19e-11 ***
## ftrtD-penicillamin 0.1533012 0.1860559 0.824 0.40997
## ftrtnot_rand -0.0067883 0.2341321 -0.029 0.97687
## fsexfemale -0.5580839 0.2285004 -2.442 0.01459 *
## fedemaappeared 0.3791800 0.2399243 1.580 0.11401
## fedemaserious 1.0097446 0.3074263 3.285 0.00102 **
## bili 0.1024013 0.0131163 7.807 5.85e-15 ***
## albumin -0.8922870 0.2027481 -4.401 1.08e-05 ***
## platelet -0.0012364 0.0008936 -1.384 0.16649
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for poisson family taken to be 1)
##
## Null deviance: 529.71 on 406 degrees of freedom
## Residual deviance: 387.11 on 398 degrees of freedom
## AIC: 715.11
##
## Number of Fisher Scoring iterations: 6Classical functions as anova, drop1,
add1 can be used for finding a suitable model:
anova(fit_cens, test = "Chisq")
drop1(fit_cens, test = "Chisq")
add1(fit_cens, scope = c("ftrt:albumin", "fedema:bili"), test = "Chisq")Find a suitable model for \(\lambda(\mathbf{Z}, \boldsymbol\beta)\)
based on pbc data and interpret it. Write also down all the
assumptions of the used probabilistic model including the formula for
\(\lambda(\mathbf{Z},
\boldsymbol\beta)\).
- Deadline for report: Monday 14th November 9:00
Maybe useful function for determining useful parametrization of continuous variables:
variables <- c("bili", "albumin", "platelet")
PlotCoefFactor <- function(variable, ngroups = 10){
# variable must be in the data
if(!is.element(variable, colnames(subdata))){stop(paste("variable", variable, "is not included in subdata!"))}
# ngroups >= 2 !!!!
if(ngroups < 2){stop("ngroups has to be at least 2!")}
othervar <- setdiff(variables, variable)
qq <- quantile(subdata[,variable], probs = seq(0,1,length.out = ngroups+1))
ff <- cut(subdata[,variable], breaks = c(-Inf, qq[2:ngroups], Inf))
model <- glm(as.formula(paste("delta ~ ftrt + fsex + fedema + ff +",
paste(setdiff(variables, variable), collapse = " + "),
" + offset(log(time))", sep = "")),
family = poisson, data = subdata)
yy <- c(0, # coefficient for the first (reference) group
model$coefficients[grep("ff", names(model$coefficients))])
xx <- (qq[1:ngroups]+qq[2:(ngroups+1)])/2
plot(yy ~ xx,
type = "b",
xlab = variable,
ylab = "Coefficient",
cex = summary(ff)/max(summary(ff))*2+0.5)
}
# For example bili
par(mar = c(4,4,1,1))
PlotCoefFactor("bili")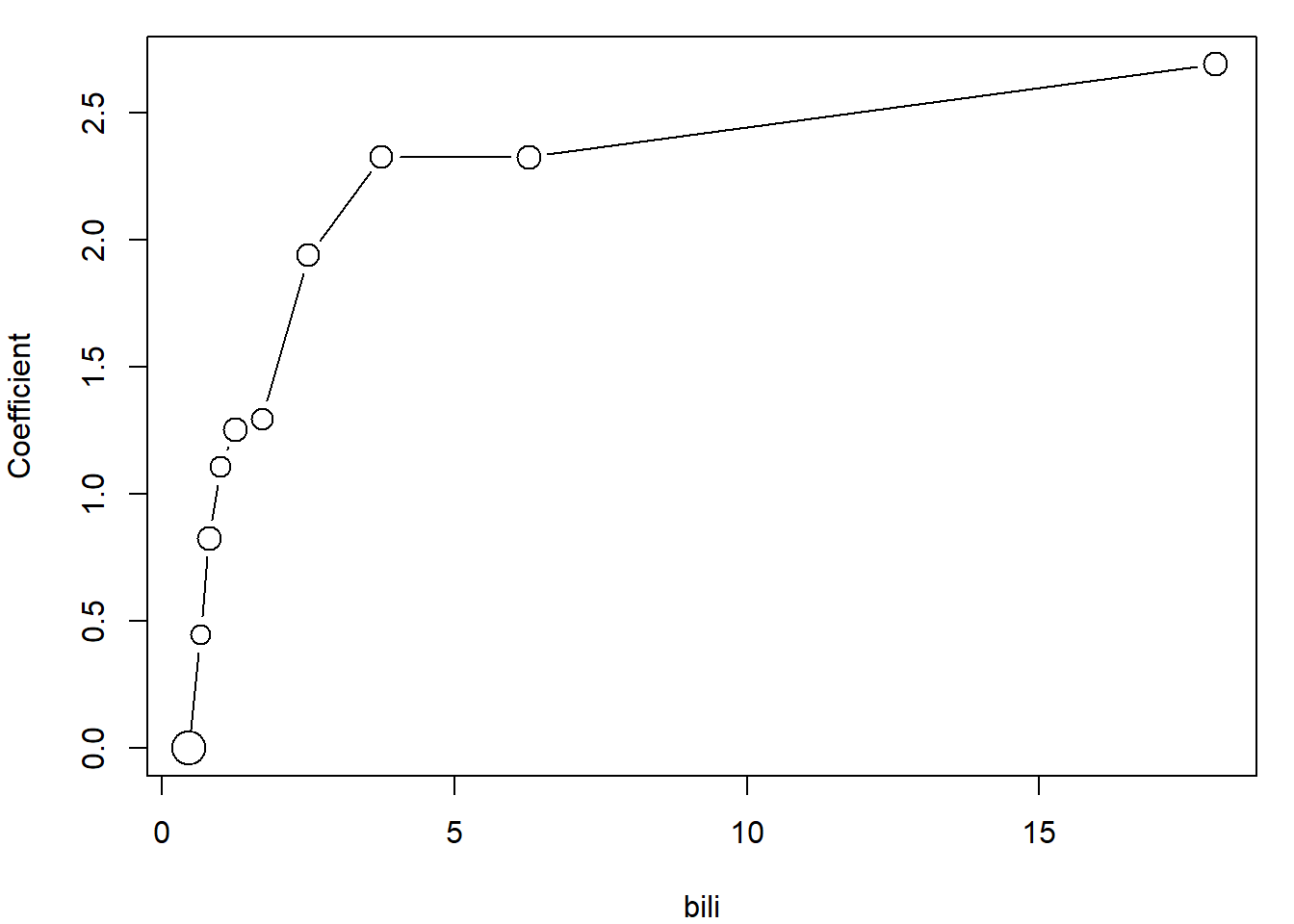
Ignoring censoring
Suppose we were not given the \(\delta\)-indicator and believe that
variable time always represents the time of death of
patient. We would still suppose the same model \[X_i = T_i | \mathbf{Z}_i \sim \mathsf{Exp}
\left(\lambda(\mathbf{Z}_i, \boldsymbol\beta) \right), \qquad
\text{where} \; \lambda(\mathbf{Z}, \boldsymbol \beta) = \lambda_0 \exp
\left\{ \boldsymbol\beta^{\mathsf{T}} \mathbf{Z} \right\}. \]
This model is exponential regression that belongs to Gamma family of GLM
and can be fitted directly using glm function in two
ways.
family = Gamma, link = "log" and
dispersion = 1
fit_exp_regr1 <- glm(time ~ ftrt + fsex + fedema + bili + albumin + platelet, family = Gamma(link = "log"), data = subdata)
summary(fit_exp_regr1, dispersion = 1)##
## Call:
## glm(formula = time ~ ftrt + fsex + fedema + bili + albumin +
## platelet, family = Gamma(link = "log"), data = subdata)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.18365 -0.42103 -0.05654 0.26064 1.58953
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 6.0701346 0.5103532 11.894 < 2e-16 ***
## ftrtD-penicillamin -0.0225165 0.1145596 -0.197 0.844181
## ftrtnot_rand -0.1896422 0.1322375 -1.434 0.151543
## fsexfemale 0.0373071 0.1624863 0.230 0.818401
## fedemaappeared -0.1099104 0.1658821 -0.663 0.507599
## fedemaserious -0.6979133 0.2587123 -2.698 0.006983 **
## bili -0.0474816 0.0122692 -3.870 0.000109 ***
## albumin 0.4358234 0.1296959 3.360 0.000778 ***
## platelet 0.0004048 0.0005244 0.772 0.440126
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for Gamma family taken to be 1)
##
## Null deviance: 199.30 on 406 degrees of freedom
## Residual deviance: 140.67 on 398 degrees of freedom
## AIC: 6709.2
##
## Number of Fisher Scoring iterations: 7Replace delta by vector of ones
fit_exp_regr2 <- glm(rep(1,dim(subdata)[1]) ~ ftrt + fsex + fedema + bili + albumin + platelet + offset(log(time)), family=poisson, data = subdata)
summary(fit_exp_regr2)##
## Call:
## glm(formula = rep(1, dim(subdata)[1]) ~ ftrt + fsex + fedema +
## bili + albumin + platelet + offset(log(time)), family = poisson,
## data = subdata)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.58954 -0.26064 0.05654 0.42103 2.18365
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -6.0701177 0.5067504 -11.979 < 2e-16 ***
## ftrtD-penicillamin 0.0225142 0.1151535 0.196 0.844990
## ftrtnot_rand 0.1896398 0.1334791 1.421 0.155391
## fsexfemale -0.0373090 0.1632920 -0.228 0.819273
## fedemaappeared 0.1099083 0.1676906 0.655 0.512195
## fedemaserious 0.6979189 0.2570672 2.715 0.006629 **
## bili 0.0474806 0.0113436 4.186 2.84e-05 ***
## albumin -0.4358263 0.1292282 -3.373 0.000745 ***
## platelet -0.0004048 0.0005343 -0.758 0.448616
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for poisson family taken to be 1)
##
## Null deviance: 199.30 on 406 degrees of freedom
## Residual deviance: 140.67 on 398 degrees of freedom
## AIC: 972.67
##
## Number of Fisher Scoring iterations: 5These approaches yield (almost) the same estimates except for change of sign. However, what is the comparison of these results to the model with censoring times?
Small simulation study
What would happen if the censoring was ignored and the parameters would be estimated based on the model \[X_i = T_i \sim \mathsf{Exp}\left(\lambda_i\right), \qquad \lambda_i = \lambda_0 \exp \left\{\beta Z_i\right\},\] however, the true model would be the one with censoring? See the script ignoring_censoring.R.