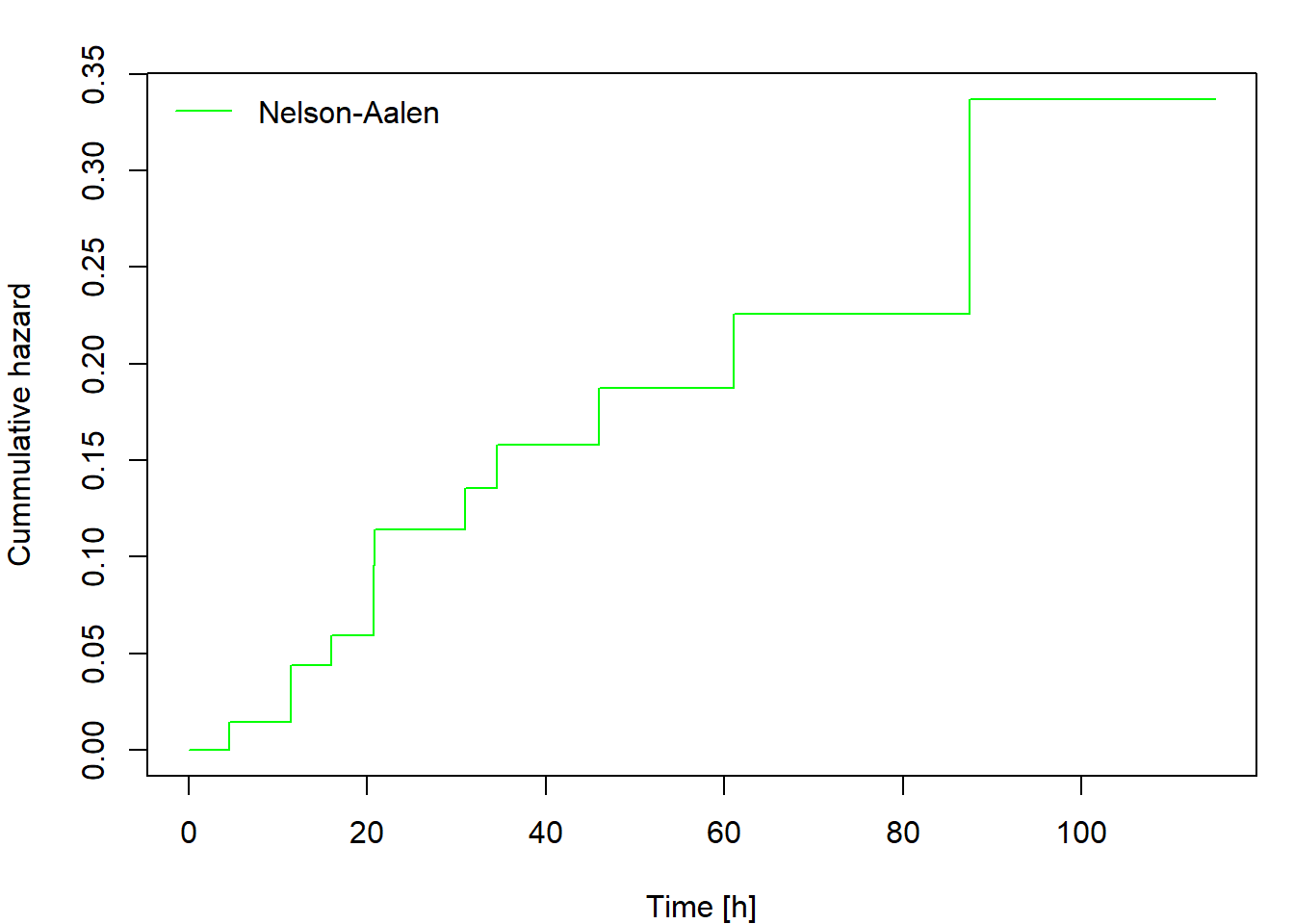
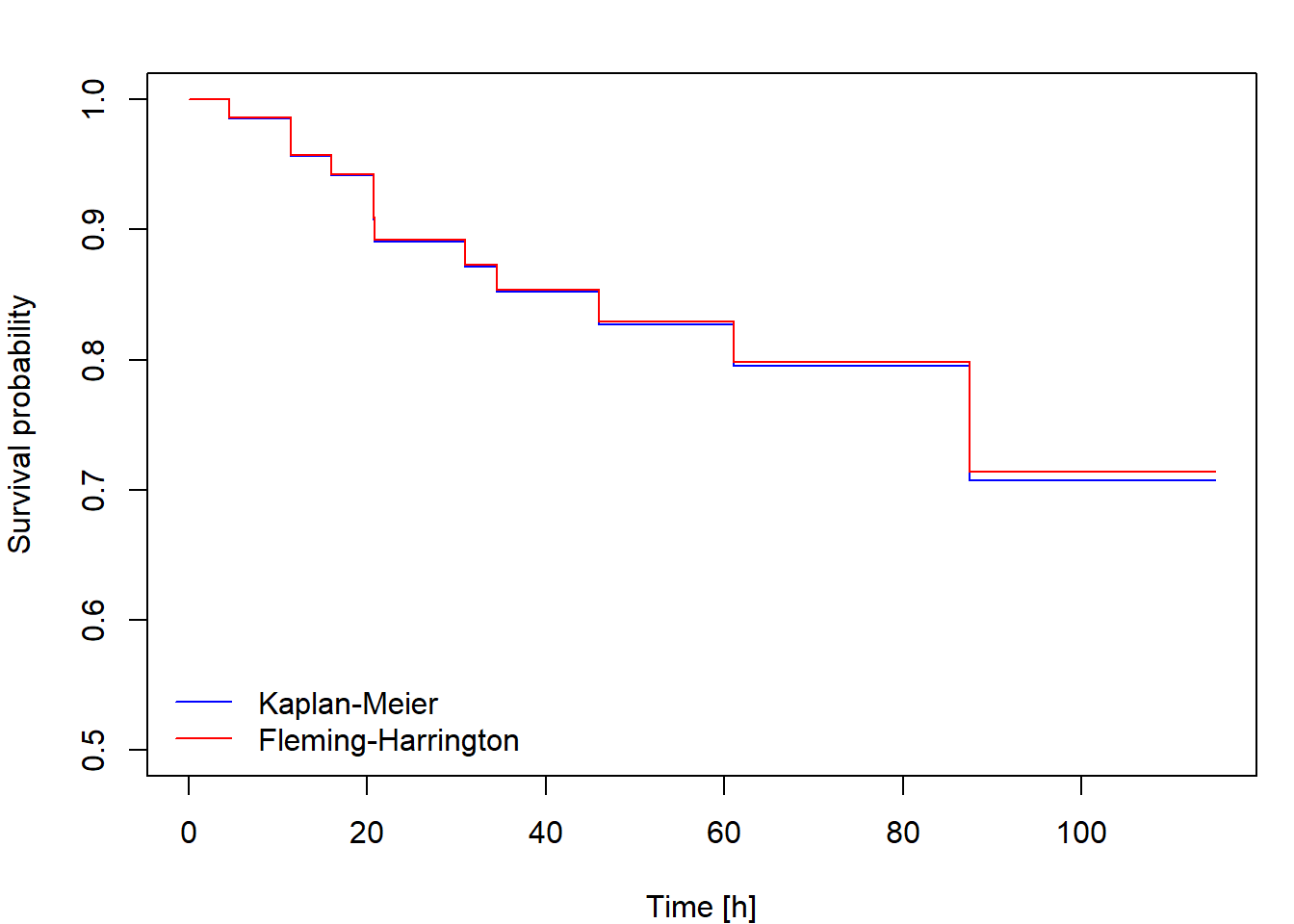
Non-parametric estimation of cumulative hazard and survival function
Jan Vávra
Exercise 2 (14th November 2022)
Lecture revision: blackboard notes.
Theory
The cumulative hazard can be estimated non-parametrically by the Nelson-Aalen estimator defined as
\[ \widehat{\Lambda}(t)=\int_0^t\frac{\,\mathrm{d} \overline{N}(u)}{\overline{Y}(u)}= \sum_{\{i: t_i\leq t\}} \frac{\Delta\overline{N}(t_i)}{\overline{Y}(t_i)}, \] where \(0<t_1<t_2<\cdots<t_d\) are ordered distinct failure times.
The Kaplan-Meier [KM] estimator of the survival function is \[ \widehat{S}(t)=\prod_{\{i: t_i\leq t\}}\biggl[1-\frac{\Delta\overline{N}(t_i)}{\overline{Y}(t_i)}\biggr]. \] If the distribution of \(T_i\) is continuous, one could define an alternative survival function estimator using the relationship \(S(t)=\exp\{-\Lambda(t)\}\) applied to the Nelson-Aalen estimator, that is \[ \widehat{S}_*(t)=\exp\{-\widehat{\Lambda}(t)\}=\prod_{\{i: t_i\leq t\}}\exp\biggl\{-\frac{\Delta\overline{N}(t_i)}{\overline{Y}(t_i)}\biggr\}. \] This is called the Fleming-Harrington [FH] estimator (sometimes also the Breslow estimator). Because \(\exp\{-h\}\approx 1-h\) for small \(h\), the difference between the KM and FH estimators is small unless the risk set size \(\overline{Y}(u)\) is not large enough, which happens at the largest failure times. The FH estimator is not suitable for discrete data, while the KM estimator works well for both discrete and continuous cases.
Understanding the formulae
The dataset fans [Nelson, Journal of Quality Technology,
1:27-52, 1969] comes from an engineering study of the time to failure of
diesel generator fans. Seventy generators were studied. For each one,
the number of hours of running time from its first being put into
service until fan failure or until the end of the study (whichever came
first) was recorded.
The data is available in the RData format from this link.
The dataframe is called fans, the variable
fans$time includes the censored failure time \(X_i\) (in hours), the variable
fans$fail is the failure indicator \(\delta_i\).
We will use this dataset to demonstrate the calculation of cumulative hazard and survival function estimates.
Using ordinary R functions (table, cumsum,
cumprod, rev, …, but not functions from the
survival library), calculate and print the following
table:
| \(i\) | \(t_i\) | \(d_i\) | \(y_i\) | \(d_i/y_i\) | \(L_i\) | \(S_i\) | \(S^*_i\) |
|---|---|---|---|---|---|---|---|
| 1 | … | … | … | … | … | … | … |
| … | … | … | … | … | … | … | … |
| d | … | … | … | … | … | … | … |
where \(i\) is the order of the failures, \(t_i\) is the ordered failure time, \(d_i=\Delta\overline{N}(t_i)\), \(y_i=\overline{Y}(t_i)\), \(L_i=\widehat{\Lambda}(t_i)\), \(S_i=\widehat{S}(t_i)\), and \(S^*_i=\widehat{S}_*(t_i)\).
Then plot the cumulative hazard estimator and both survival function estimators. Include in your report the code for calculating and plotting the results and the output you obtained.
Calculating the estimators using library(survival)
The basic functions for censored data analysis are available in R
library survival. Censored data are entered as special
survival objects created in this way:
library(survival)
Surv(time,del)
## see help(Surv)
# used as a response variable in a model formula
# right censoring is set as defaultwhere time is a numeric vector containing the censored
failure times and del is a numeric vector containing
failure indicators (1 = failure, 0 = censored). Survival objects are
used as input to functions providing analysis of censored data.
Survival function estimates are calculated by the function
survfit. The most rudimentary use of this function is
fit <- survfit(Surv(time,del)~1, data=data)This calculates the KM estimator. The FH estimator is obtained by
adding an option type
fit <- survfit(Surv(time,del)~1, type="fleming", data=data)The results can be plotted easily by calling
plot(fit)For demonstration, we will use the dataset aml included
in library(survival). Survival in patients with Acute
Myelogenous Leukemia. The question at the time was whether the standard
course of chemotherapy should be extended (‘maintenance’) for additional
cycles.
print(aml)## time status x
## 1 9 1 Maintained
## 2 13 1 Maintained
## 3 13 0 Maintained
## 4 18 1 Maintained
## 5 23 1 Maintained
## 6 28 0 Maintained
## 7 31 1 Maintained
## 8 34 1 Maintained
## 9 45 0 Maintained
## 10 48 1 Maintained
## 11 161 0 Maintained
## 12 5 1 Nonmaintained
## 13 5 1 Nonmaintained
## 14 8 1 Nonmaintained
## 15 8 1 Nonmaintained
## 16 12 1 Nonmaintained
## 17 16 0 Nonmaintained
## 18 23 1 Nonmaintained
## 19 27 1 Nonmaintained
## 20 30 1 Nonmaintained
## 21 33 1 Nonmaintained
## 22 43 1 Nonmaintained
## 23 45 1 NonmaintainedWe get a full description of our estimates of survival function by
summary applied to survfit object:
fit_KM <- survfit(Surv(time,status)~1, data=aml)
summary(fit_KM)## Call: survfit(formula = Surv(time, status) ~ 1, data = aml)
##
## time n.risk n.event survival std.err lower 95% CI upper 95% CI
## 5 23 2 0.9130 0.0588 0.8049 1.000
## 8 21 2 0.8261 0.0790 0.6848 0.996
## 9 19 1 0.7826 0.0860 0.6310 0.971
## 12 18 1 0.7391 0.0916 0.5798 0.942
## 13 17 1 0.6957 0.0959 0.5309 0.912
## 18 14 1 0.6460 0.1011 0.4753 0.878
## 23 13 2 0.5466 0.1073 0.3721 0.803
## 27 11 1 0.4969 0.1084 0.3240 0.762
## 30 9 1 0.4417 0.1095 0.2717 0.718
## 31 8 1 0.3865 0.1089 0.2225 0.671
## 33 7 1 0.3313 0.1064 0.1765 0.622
## 34 6 1 0.2761 0.1020 0.1338 0.569
## 43 5 1 0.2208 0.0954 0.0947 0.515
## 45 4 1 0.1656 0.0860 0.0598 0.458
## 48 2 1 0.0828 0.0727 0.0148 0.462We calculate and plot KM and FH estimates like this
fit_FH <- survfit(Surv(time,status)~1, type="fleming", data=aml)
par(mar=c(4,4,1,1))
plot(fit_KM, conf.int=FALSE, col="blue",
xlab = "Time", ylab = "Survival probability")
lines(fit_FH, conf.int=FALSE, col="red")
legend("topright", c("Kaplan-Meier", "Fleming-Harrington"),
col = c("blue", "red"), lty = 1, bty = "n")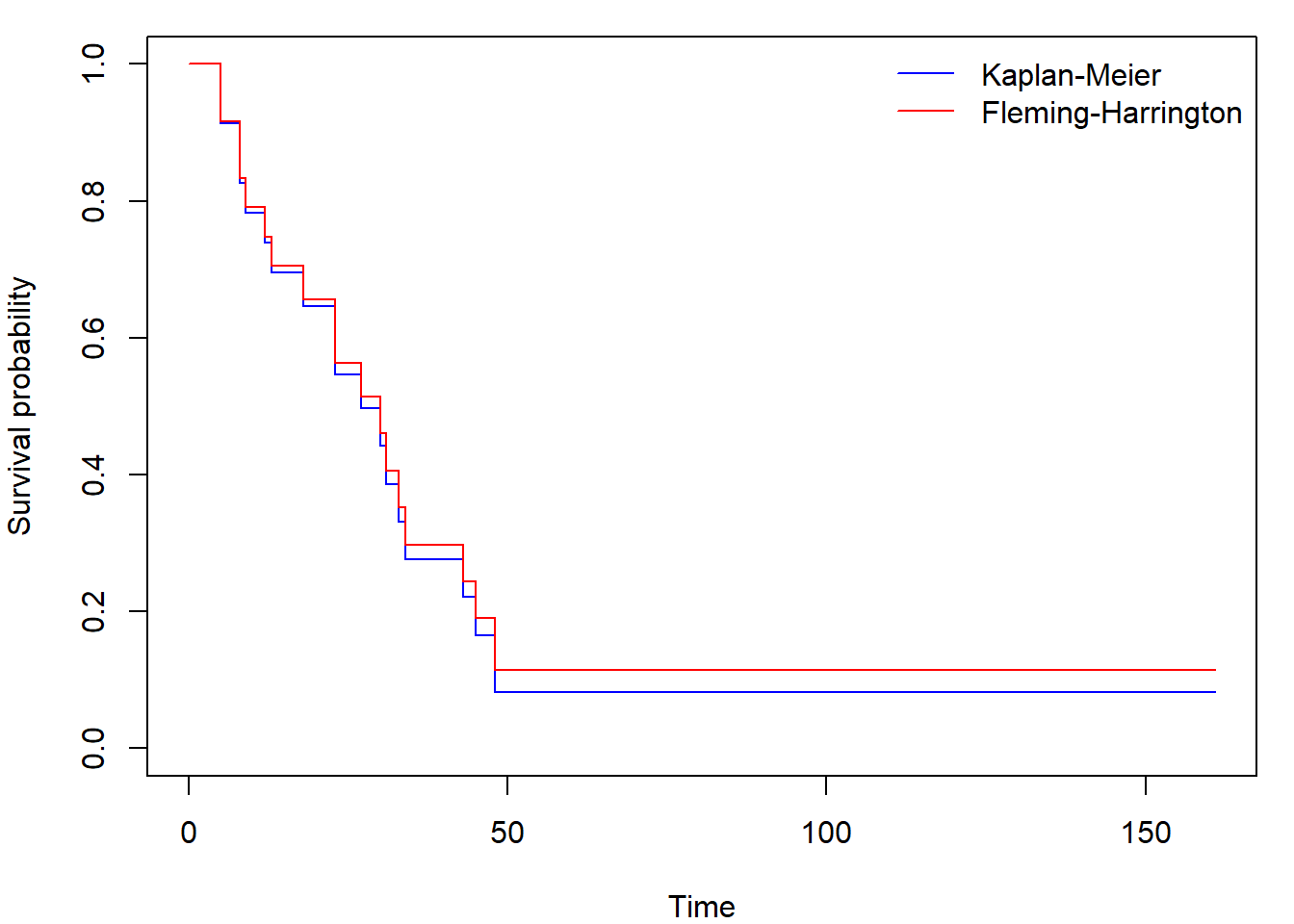
We suppressed the plotting of the confidence intervals.
Sometimes you can come across figures with crosses at times where an
observation was censored (set mark.time = T, default
mark is +):
par(mar=c(4,4,1,1))
plot(fit_KM, conf.int=FALSE, col="blue", mark.time = T,
xlab = "Time", ylab = "Survival probability")
legend("topright","Kaplan-Meier",
col = "blue", lty = 1, bty = "n")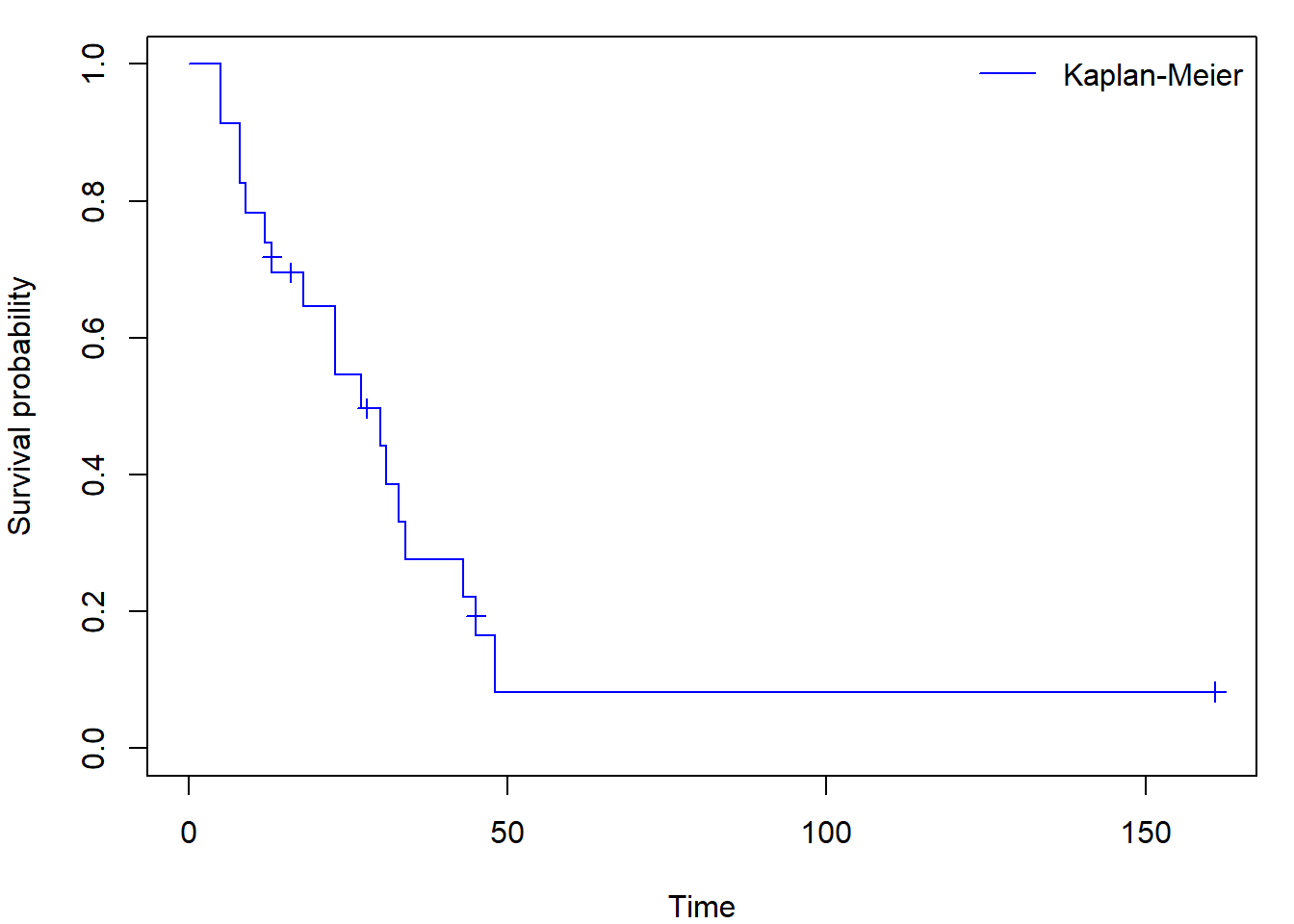
Using the formula notation, we can easily create and plot separate
curves for two distinct subgroups of observations (declared by factor
variable x within model formula instead of
1):
fit_gr <- survfit(Surv(time,status) ~ x, data=aml)
par(mar = c(4,4,1,1))
plot(fit_gr, col=c("green","magenta"), xlab = "Time", ylab = "Survival probability")
legend("topright", levels(aml$x), col = c("green","magenta"), lty = 1, bty = "n")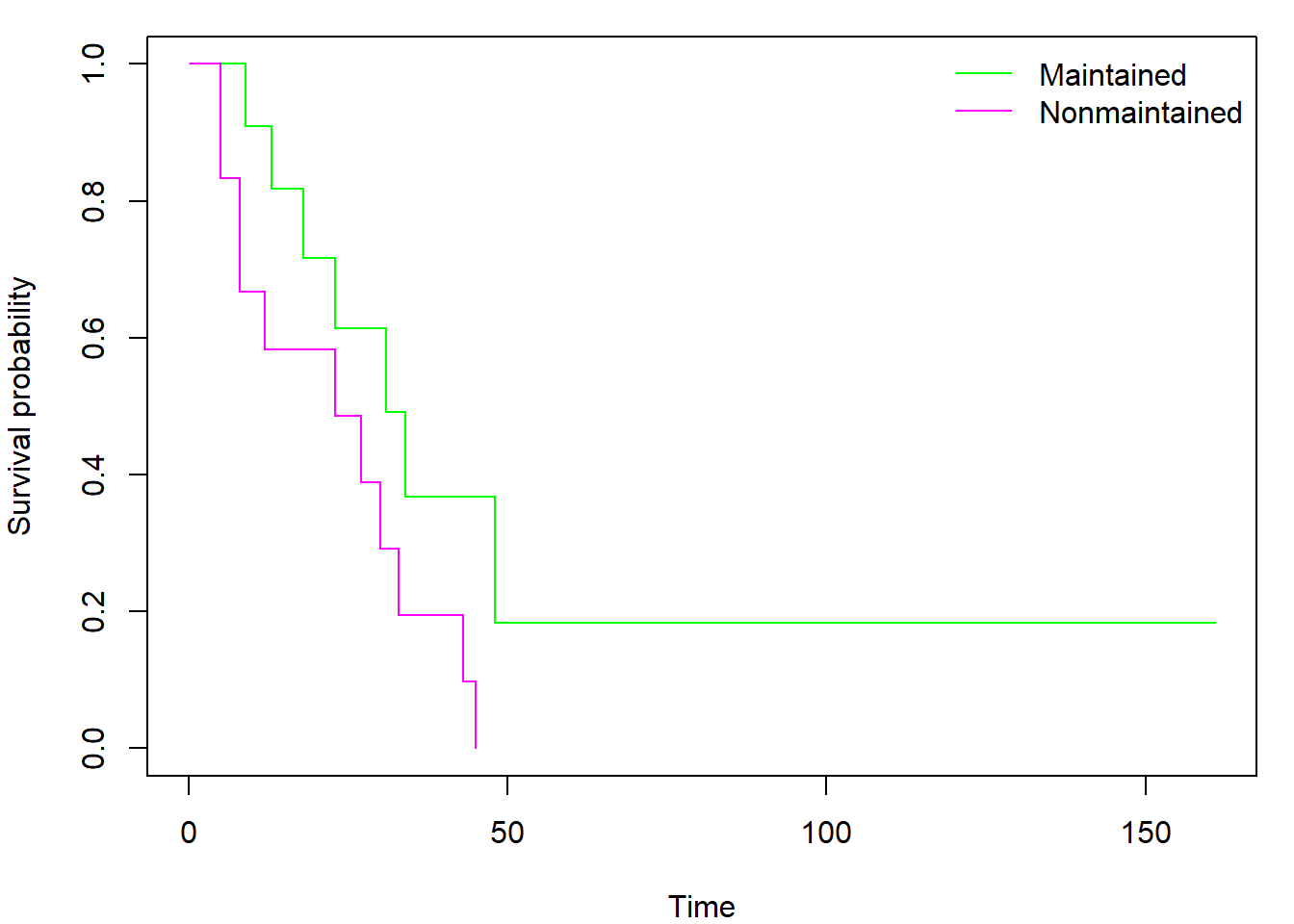
It is interesting to investigate the contents of the
survfit object by calling str(fit_KM). By
doing so, you may discover also the Nelson-Aalen estimator of cumulative
hazard function hidden in $cumhaz. Regardless of selected
type (kaplan or fleming)
$cumhaz always contains Nelson-Aalen estimator. If you need
cumulative hazard estimator derived from Kaplan-Meier estimator of
survival function you have to use the transformation \(\Lambda(t) = - \log \left( S(t) \right)\).
You can also easily plot the cumulative hazard function(s) by adding
fun = "cumhaz" into the plot command (see
help(plot.survfit) for other uses of fun
parameter):
par(mar = c(4,4,1,1))
plot(fit_gr, col=c("green","magenta"), xlab = "Time", ylab = "Cumulative hazard function",
fun = "cumhaz")
legend("topright", levels(aml$x), col = c("green","magenta"), lty = 1, bty = "n")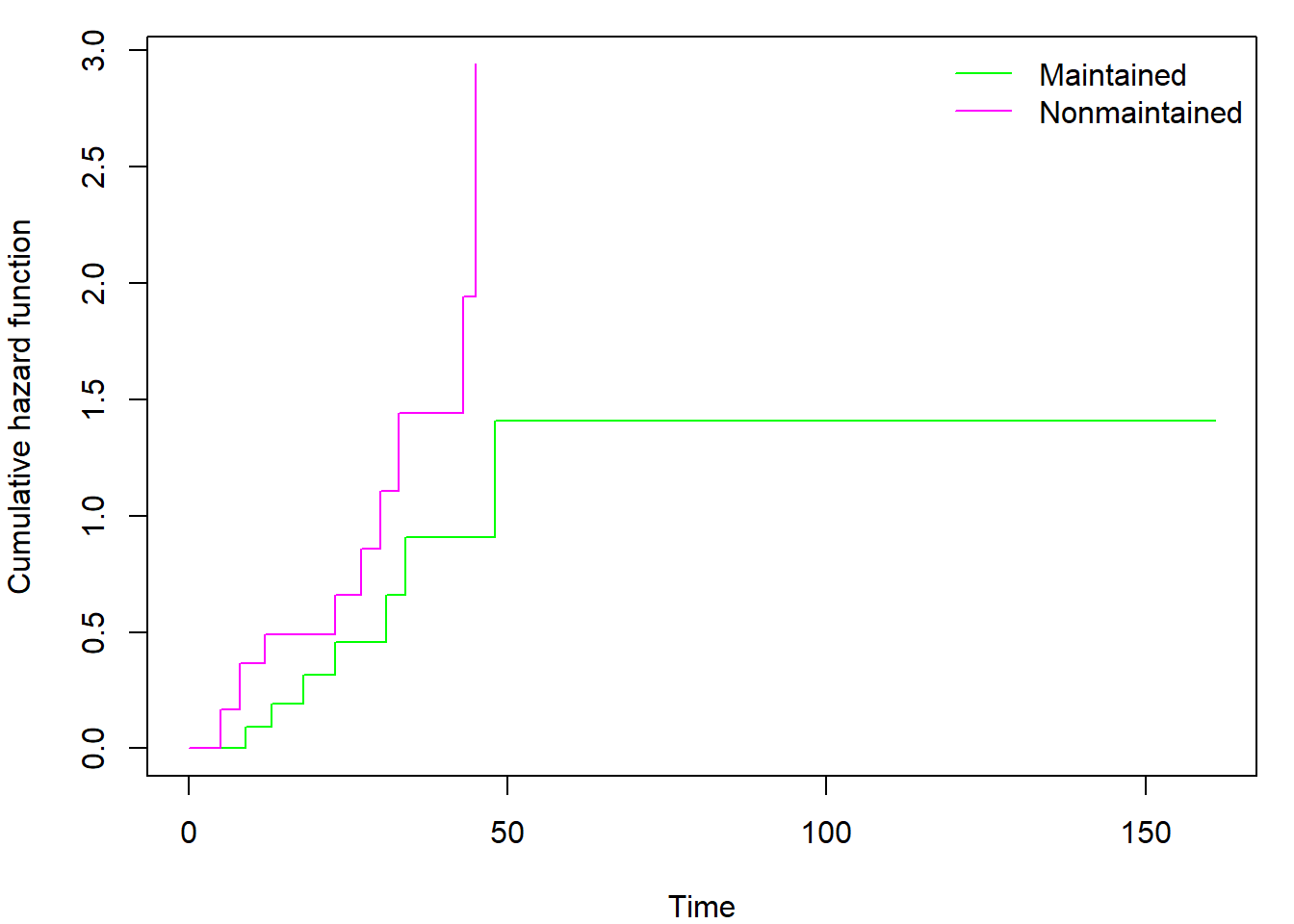
You can also use library(ggplot2) in cooperation with
library(survminer) to obtain nicer plots:
library(survminer)
library(ggplot2)
ggsurvplot(fit_gr, conf.int = F, censor= T,
ggtheme = theme_minimal())
Calculate and plot the cumulative hazard estimator and both survival
function estimators for fans data using the function
survfit. Compare the results to those you obtained in Task
1. Include the code, output and comments in your report.
Advice: Implement your own function for creating nicely looking step-functions. You will surely use it in the future.
- Deadline for report: Monday 21st November 9:00
Show that the Nelson-Aalen estimator satisfies the equation \[ \sum_{i=1}^n \widehat{\Lambda}(X_i)=\sum_{i=1}^n \delta_i. \]
Show that under absence of censored data the Kaplan-Meier estimator becomes classical empirical estimator: \[ \widehat{S}(t) = \prod\limits_{\{j: t_j \leq t\}} \left[1 - \dfrac{\Delta\overline{N}(t_j)}{\overline{Y}(t_j)}\right] = \frac{1}{n}\sum \limits_{i=1}^n \mathbb{I}(X_i > t), \quad \forall t >0. \]